January 10, 2025
【Case study】Development of a large-scale virtual screening drug discovery platform (XEDOCK) based on Kubernetes
Development of a large-scale virtual screening drug discovery platform (XEDOCK) based on Kubernetes
In response to the increasingly high cost of drug research and development, the technique of virtual screening, which uses computational modeling to select potential drug candidates from large collections of compounds such as commercial and virtual compound libraries, is expected to improve the efficiency of drug discovery research. Improvements are expected in terms of the cost, speed and safety of drugs discovered, due to a significant reduction in the number of real-world experiments required, such as compound synthesis and biological testing, and the more effective identification of ideal drug candidates for development.
1.Platform for virtual screening of ultra-large scale compound libraries
Although virtual libraries are sometimes created/selected semi-manually by humans, most often we use pre-existing compound libraries, such as those prepared internally by each pharmaceutical company, public libraries such as ChEMBL and PubChem, or commercial compound libraries provided by reagent suppliers. In recent years, the available libraries have become so large (billions to trillions of compounds) that a new platform was needed to efficiently handle the large data sets involved.
Activity prediction of compounds from such data sets generally uses one of two methods: the application of statistical methods based on existing structure-activity relationship data, or structure-based prediction methods such as molecular docking. In either case, the increasingly large data sets involved require the use of more memory, storage, and computational resources (CPU/GPU), thus it becomes necessary to allocate these resources appropriately.
1.1.A Kubernetes-based virtual screening platform
Kubernetes is a scalable, container-based orchestration platform for managing containerized applications on a large scale. Originally developed by Google, Kubernetes is now being developed as a fundamental platform by the Cloud Native Computing Foundation, and many public cloud services (AWS, Azure, GCP, etc.) provide Kubernetes as one of their features. Thus, it can be used in a wide variety of environments. In addition, since each microservice is enclosed in a separate container, it has high availability, automatically re-running even if there are failures due to non-reproducible bugs, resource shortages, or sudden shutdowns due to preemption.
Xeureka has developed the XEDOCK system, a microservice-based system based on Kubernetes as a platform for the virtual screening of large compound libraries. This system is such that docking is distributed, processed, and executed with a single click by uploading a simple configuration file. The system dynamically scales computing resources according to the computing load based on the size and type of tasks loaded into the queue, so that a large number of virtual machines (VMs) are reserved for high loads and a minimal number of VMs are reserved for low loads.
In the XEDOCK system, the head node sends tasks to a queue according to the manifest, and each microservice receives tasks from the queue and processes them. In addition, apart from simply executing a large number of tasks, the queue can be monitored and depending on the progress of the next task can be connected to the next separate process.
All workers are implemented in such a way that they can be resumed, so that even if there is an irregular shutdown caused by a temporary hardware failure or a spot instance preemption or memory leak, the overall task will continue to progress as it is restarted.
This mechanism is suited for executing a large number of tasks in parallel, for example, molecular docking or building machine learning models.
2.Example of XEDOCK system in action
This section illustrates docking with AutoDock-Vina as an example.
2.1.Upload molecular pool
Molecular pool formats include SDF, CSV, etc.
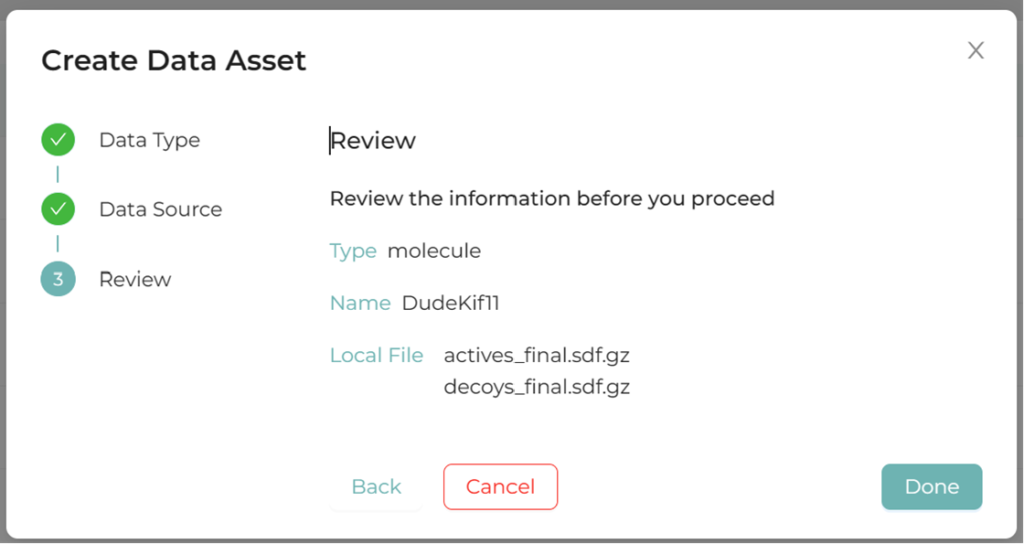
Figure 1. Molecular pool registration screen in XEDOCK
2.2.Upload receptor
Upload a PDB or PDBQT file.
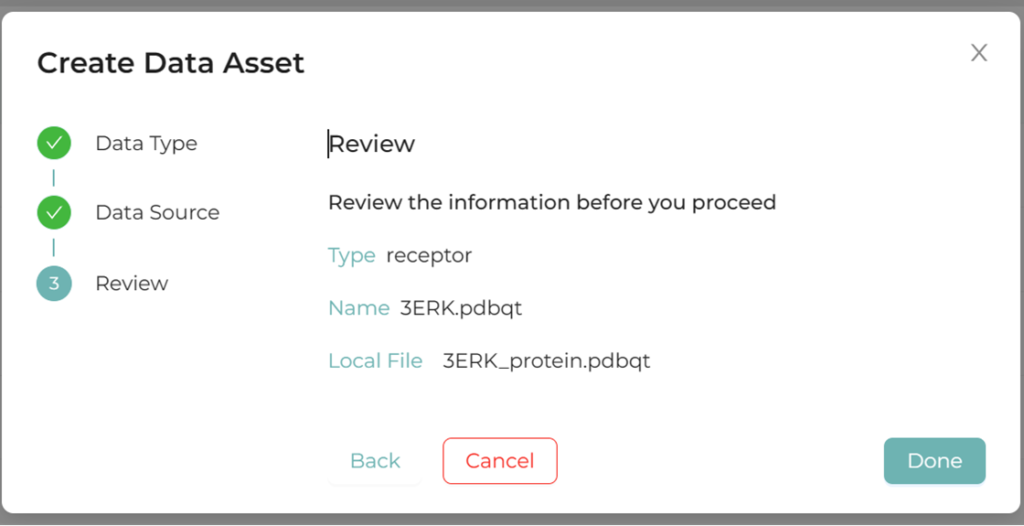
Figure 2. Receptor registration screen in XEDOCK
2.3.Upload experimental conditions
The manifest file for docking conditions is shown below.
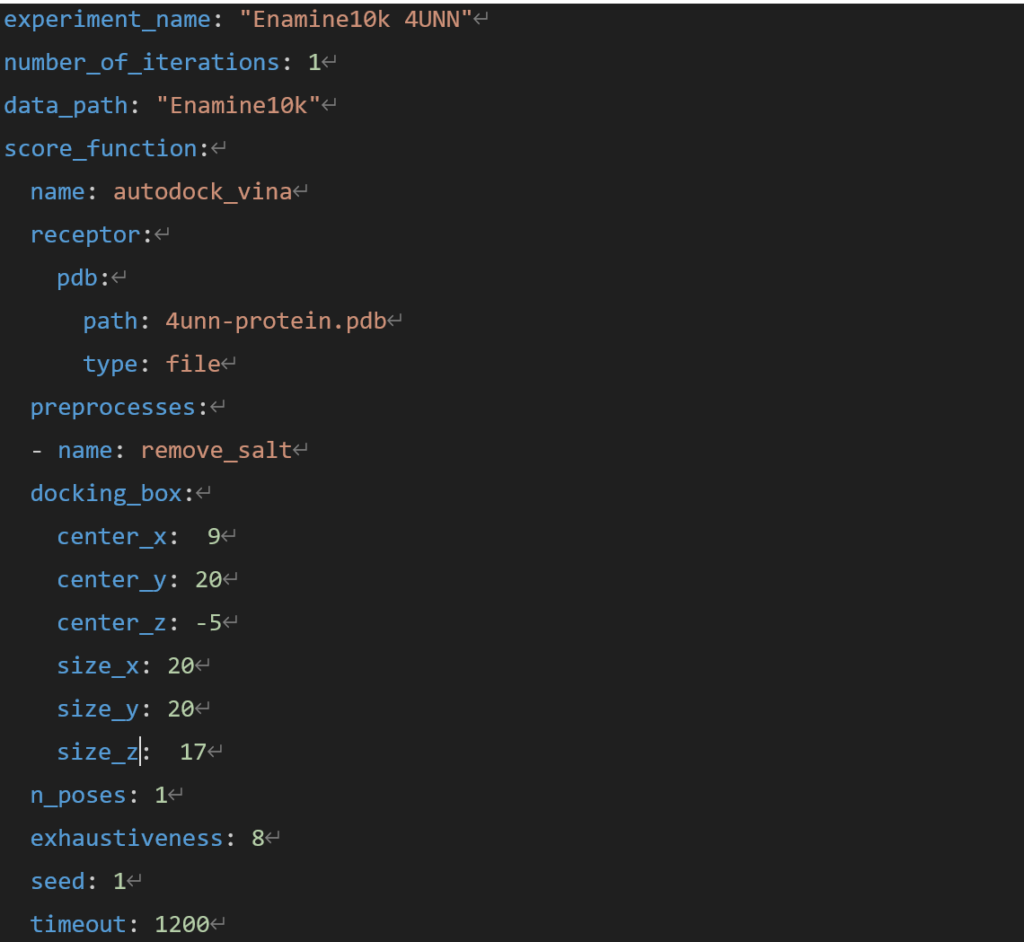
Figure 3. Example of XEDOCK manifest file
Figure 4. Manifest file registration screen in XEDOCK
Once docking is initiated in this manner, AutoDock-Vina will secure computing resources from the cloud, generate conformations from SMILES or MolBlocks contained in the uploaded molecular pool file, and convert them to a format that AutoDock-Vina can interpret (PDBQT). The docking of 10,000 compounds was completed in 25 minutes on Microsoft Azure (F16-v2 × 50 (spot instance)).
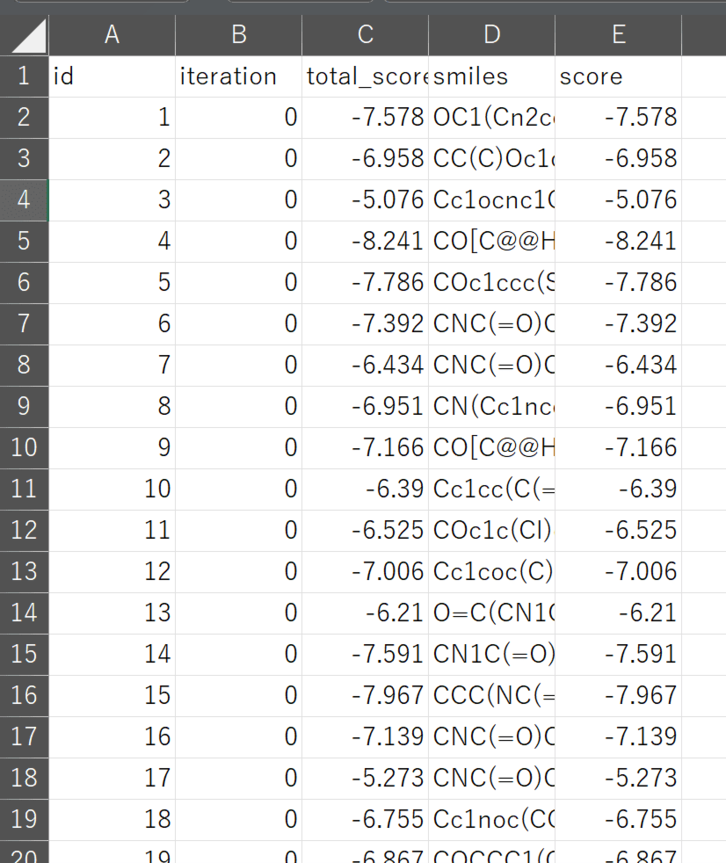
Output from the run is a CSV file containing a summary of the scores and 10,000 docked poses in SDF format.
2.4.Summary of results
Figure 5. Example of XEDOCK output file
 Figure 6. Example of docking poses from XEDOCK output SDF file
Figure 6. Example of docking poses from XEDOCK output SDF file
Computational resources allocated during computation are released through AKS (Azure Kubernetes Service) when docking is completed.
2.5.Application example: docking + active learning
A more complex application example is the use of pool-based active learning to perform virtual screening.
Active learning can be used to select an optimal subset of the full, very large data set in order to most effectively train a model. It is a particularly effective technique to use when labeling data is costly.
Molecular docking is a computationally demanding task that can be described as costly in terms of data labeling. Therefore, it is considered to be a suitable process for the use of active learning.
The XEDOCK system can handle light to heavy computations, from small data sets to data sets with billions of records. It is also equipped with a variety of active learning algorithms, prediction models, compound features, and scoring functions that can be used in combination.
Furthermore, since there is no feature or prediction model that can be expected to handle all kinds of problems, the system automatically selects the function or ensemble of functions to provide the best model for any situation.
Details of the active learning process are as follows.
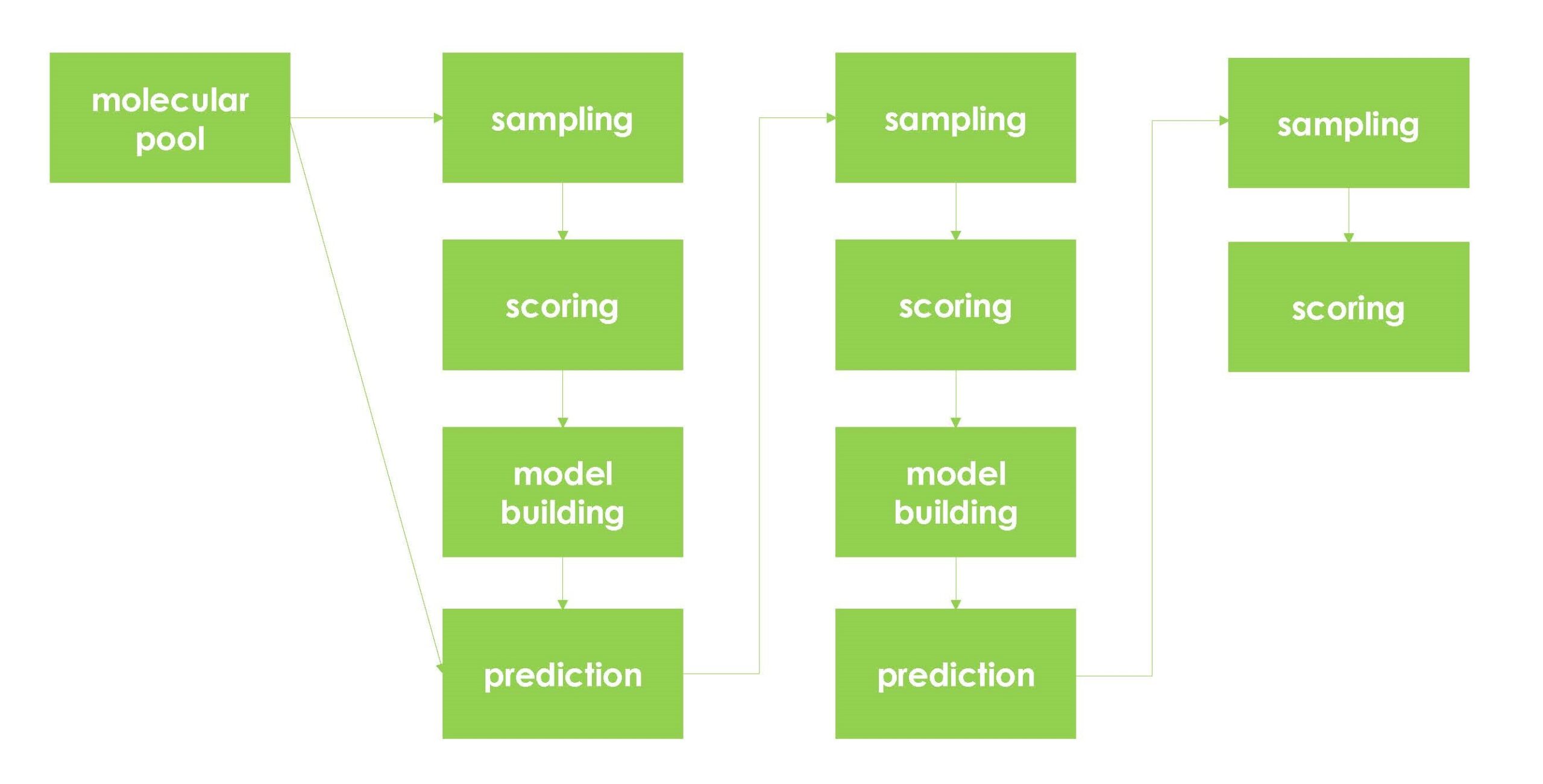
Figure 7. Active learning flow
Active learning based on molecular docking requires four functionally distinct microservices (sampling, labeling, model building, and prediction), as well as a service (head node) to control and monitor them.
While such a workflow can be looped into a single process, Xeureka has built this as a microservice on a cloud service. The XEDOCK system can be easily deployed on a variety of cloud platforms and on-premise environments. In the previous example, the system was implemented on Azure, but it can also be deployed on AWS, for example, as shown below.
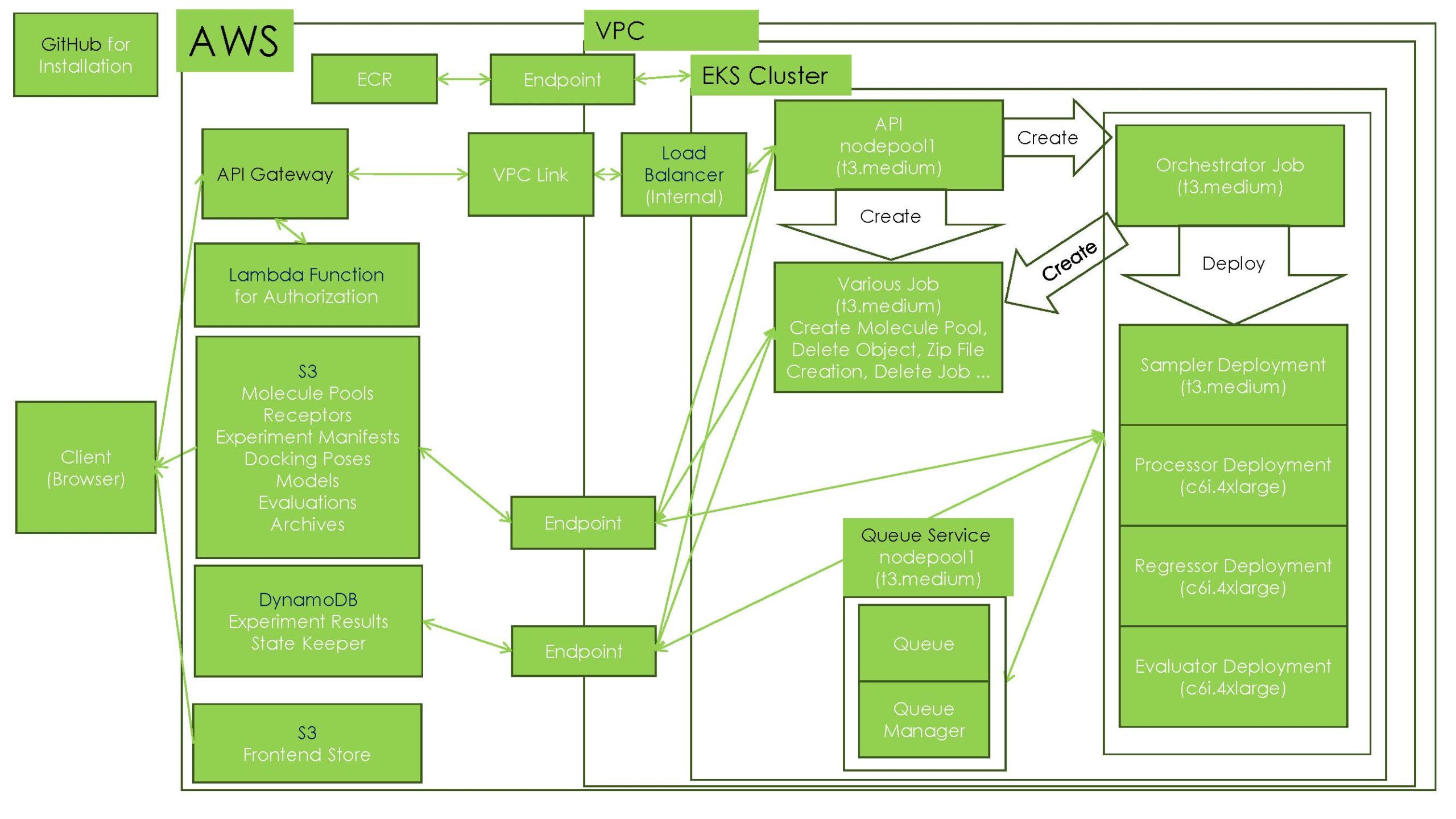
Figure 8. Example of XEDOCK configuration on AWS
When the API that initiates an active learning experiment is called, the orchestrator job is launched and sends messages to the queue service with the necessary tasks. The sampler, processor, regressor, and evaluator microservices are scaled according to the state of the queue, each receiving messages from the queue and processing tasks. The orchestrator controls the flow of active learning while monitoring the entire process.
3.Active learning performance
Active learning is typically aimed at maximizing the performance of the model being built, but when used in virtual screening, it also serves the purpose of sampling and labeling the more interesting compounds in the active learning loop. In other words, while it is important to give accurate predictions, it is also important to identify the more interesting compounds quickly.
The XEDOCK system supports a variety of compound features, predictive models, scoring functions (e.g., docking software), and sampling. For features and predictive models, multiple models can be trained before selecting the one with the best performance, or an ensemble of multiple models can be used.
 Figure 9. Prediction models and sampling methods available in XEDOCK.
Figure 9. Prediction models and sampling methods available in XEDOCK.

Figure 10. Examples of scoring functions available in XEDOCK
3.1.Example of active learning with the XEDOCK system
One well-studied benchmark data set for active learning in docking is a set of 100 million compounds docked with DOCK against AmpC (Reference). In the example below, we refer to this data set and measure the performance index for searching the top 1000 compounds with the highest DOCK docking score out of the total of 100 million compounds in the data set.
In recent years, deep learning-based methods have also shown their ability to predict compound properties, and the XEDOCK system also supports multi-layer perceptrons for features and MPNNs that focus on the graph structure of structural formulas. In the sampling of 100,000 compounds over 6 repetitions, XEDOCK has shown enrichment of about 70% and about 100-fold.
Furthermore, we have found that our protocol performs better when the number of cycles is increased, even when using a computationally inexpensive method such as simple fingerprinting plus a classical learning model. Specifically, we repeated the evaluation of 10,000 compounds per loop, which is 1/10 of the previous example. This method yielded about 70% of the desired compounds after 30 cycles, and about 80% at 60 cycles. In other words, by looking at 0.6% of the total data set, we were able to capture 80% of the top 0.001%, which is 1/100 of the compounds to be labeled.
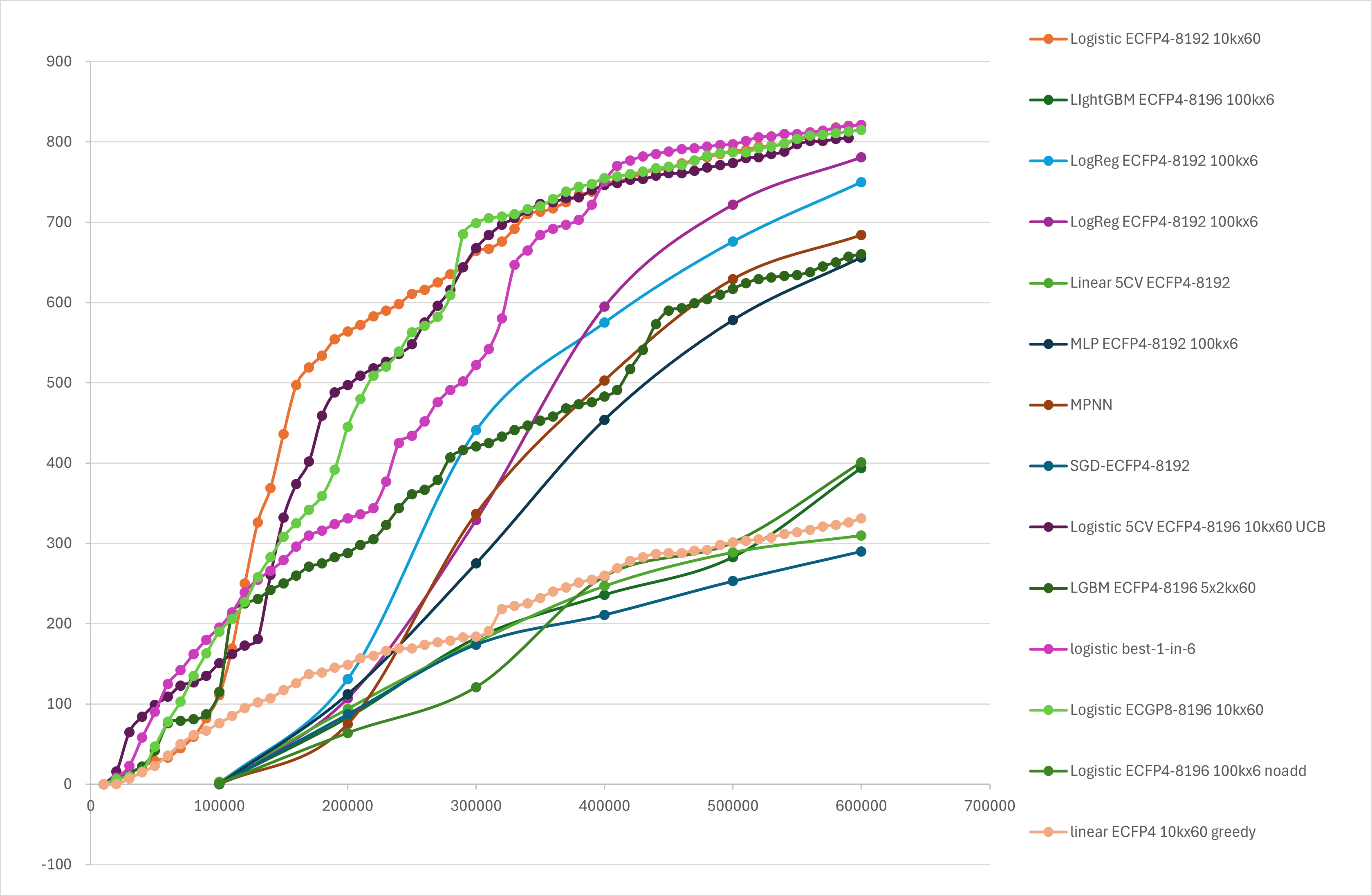
Figure 11. Comparison of active learning for docking scores using XEDOCK
The computational resources required for this were about 3 hours on a single node of Azure F16-v2 VM (16 CPUs). For larger data sets, a higher-performance VM should be sufficient.
In this blog, we have presented the example of a virtual screening study conducted by docking simulation of small molecule compounds, but the applications are not limited to small molecules or to docking methods. Additional input/output interfaces can by implemented for other objects, scoring methods or applications, and predictive models can be generated for physical properties, biological activity and more.
Summary
In this column, we have introduced the XEDOCK system developed and marketed independently by Xeureka. The system can be expanded to include scoring and sampling methods other than docking upon request. We can also handle new modalities – the system is not limited to low-molecular compounds. If you are interested in our services, please feel free to contact us from the contact page below!