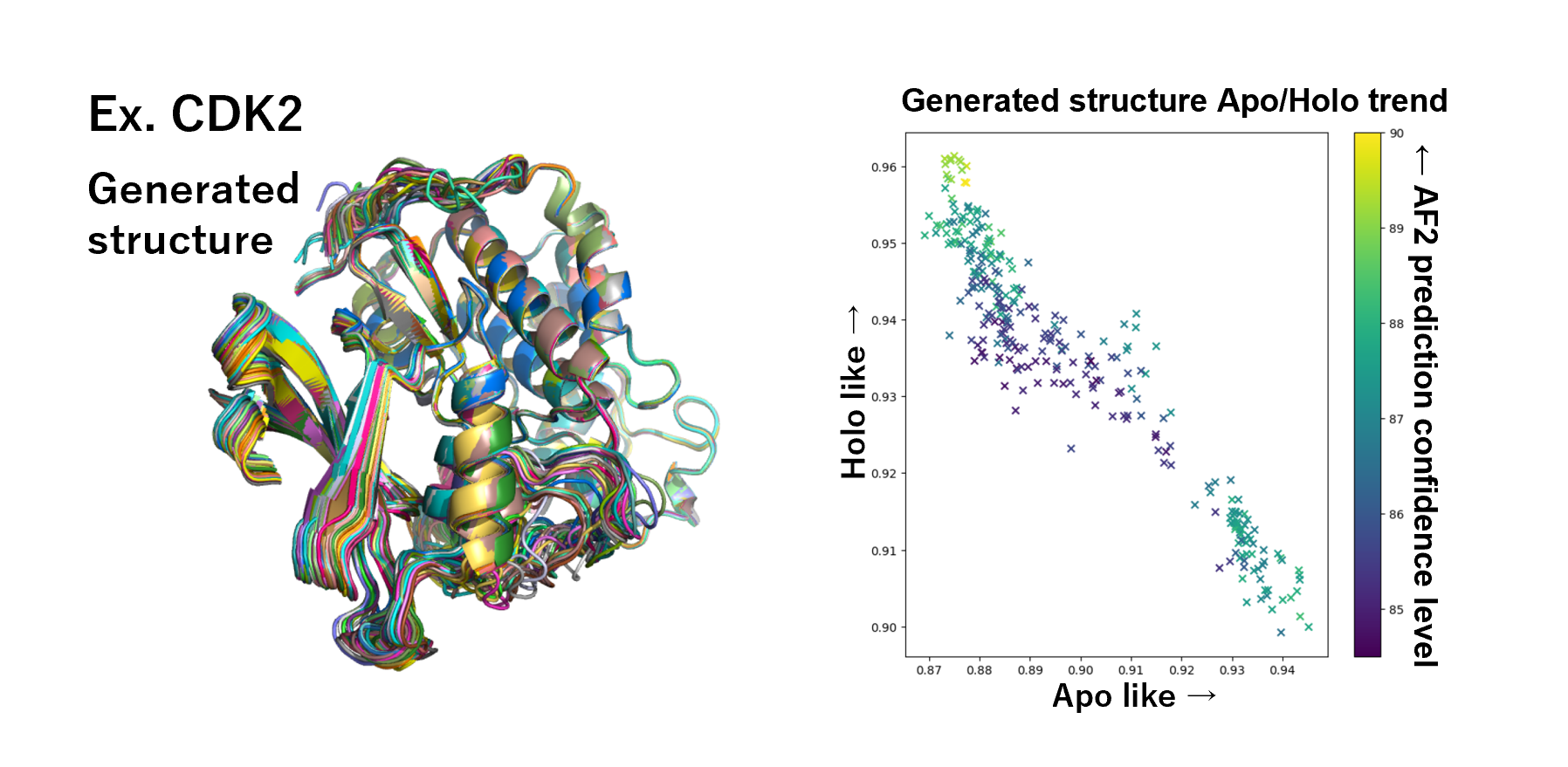
To conduct SBVS with a probability of success, it is important to use not only the 3D structure of the target protein alone (Apo structure) but rather the complex between the target protein and a ligand (Holo structure). However, high-quality complex structures are often unavailable. Therefore, we have developed and implemented a structural optimization method to derive the Holo structure from the Apo structure, as illustrated below.
Optimization of structure prediction parameters using AlphaFold2
AF2 is a prediction model trained on structural data from multiple Apo structures, so it typically generates predicted structures that are Apo structures or close to them.
However, by adjusting the structure prediction parameters of AF2, it is possible to generate structures close to the Holo protein.
Develop a method to select from among the generated structures for models that most closely match the target structure.
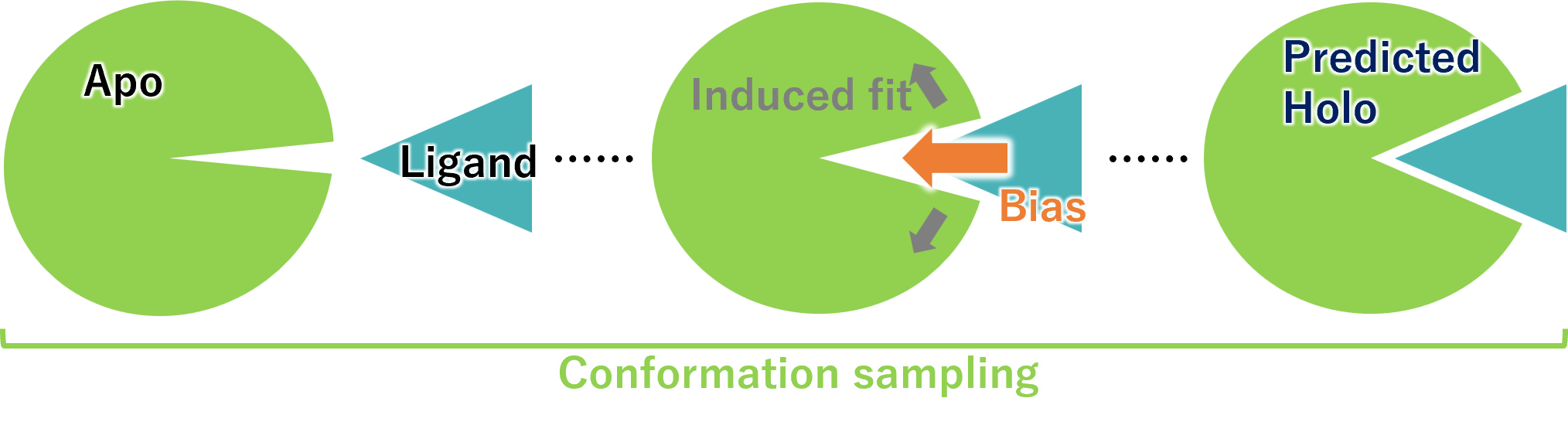
Holo structure prediction by molecular dynamics simulation
Using the structure of the Apo protein and known ligands, ligands are inserted into the protein by simulation.
Comprehensive structural sampling to obtain 3D structures of complexes close to the Holo protein.